The Tien Group
Boston University
Department of Biomedical Engineering
44 Cummington Mall
Boston, MA 02215
We are an interdisciplinary group of
researchers who invent new types of biomaterials, for uses that range from
basic studies of quantitative physiology to clinical applications in
regenerative medicine. Of particular
interest are perfusable, microfluidic materials whose internal geometries mimic
the organization of native vascular networks.
With these materials, we seek to solve one of the long-standing
challenges in tissue engineering: how to form clinically relevant volumes of
tissue that are nourished and drained by functional microvessels. We are also interested in developing
vascularized microphysiological systems for studying vessel-tissue interactions
in vitro, particularly in the context of cancer progression. We are currently in our 24th year
at Boston University.
Our research addresses the following
questions:
·
How
does one synthesize and vascularize perfusable biomaterials?
·
What
principles govern vascularization of biomaterials? Can one distill these principles into a
computer algorithm for rational scaffold design?
·
How
does one scale up vascularized materials to clinically relevant sizes? How do such materials behave upon perfusion
in vivo?
To answer these questions, we develop
unconventional methods to organize vascular and non-vascular cells and
extracellular components into perfused, micropatterned tissues. We use traditional techniques of microvascular
physiology (along with a healthy mixture of ideas from vascular cell biology,
transport phenomena, biomechanics, and numerical modeling) to analyze, predict,
and control the behavior of these tissues.
Below, we invite you to read about the group
and its research interests, publications, and resources. For further information, please contact us
directly.
GROUP INFORMATION
Principal
Investigator: Joe Tien
Address: We are located on the
7th floor of the Engineering Research Building (44 Cummington Mall), in rooms
713, 715, and 717. [Map]
Phones: (617) 358-3055 [Joe’s
office—ERB 717]
(617) 358-2831 [Main lab—ERB
715]
(617) 353-5557 [Lounge—ERB
713]
Fax: (617) 358-2835
RESEARCH INTERESTS
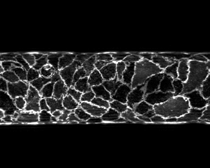 Mechanics
of vascularization
Mechanics
of vascularization
Much
of our work is focused on understanding how vascularization of microfluidic
scaffolds occurs and what factors control its long-term success. We have found that the microfluidic structure
of a scaffold is important in controlling the initial formation of open
vessels, but it is insufficient to guarantee sustained perfusion. Through a combination of experimental and
computational studies, we have shown that mechanical stresses at the cell-scaffold
interface determine whether the vascular lining remains adherent or detaches
over time. We have begun to apply this
physical theory of vascularization to a variety of challenging problems,
including the vascularization of capillary-scale channels and the formation of
functional lymphatic microvessels in which the mechanical stresses are
inherently destabilizing.
 Techniques for patterning biological
materials
Techniques for patterning biological
materials
We
have a long-standing interest in inventing new techniques for patterning
biological materials. In fact, our work
on vascularization is based on subtractive methods that we developed in-house
to create single channels and entire networks within hydrogels of extracellular
matrix. We are always interested in
patterning scaffolds with ever finer resolution, greater three-dimensional
connectivity, and larger network sizes.
Current efforts are focused on specific applications, including the
development of valves that can actively pump fluids and the creation of
large-scale microfluidic scaffolds suitable for composite tissue engineering.
Engineering
vascularized microphysiological systems
Our
latest work is geared towards developing microphysiological systems
("organs-on-a-chip") that model the interactions of vessels and
non-vascular tissue in human disease. We
have created microfluidic models of human breast cancer and its progression
towards vascular invasion, an obligate step in metastasis. These models provide a well-controlled system
for testing whether a particular element of the tumor microenvironment, such as
interstitial flow or the presence of adipose cells, alters the rate at which
tumor cells escape into a nearby vessel.
We have also begun to develop vascularized models of human obesity,
which can be used to understand the interplay between breast cancer, obesity,
and vascular invasion.
MEMBERS (Current in bold)
|
|
Joe Tien |
jtien
| bu_edu |
Principal
investigator |
|
|||
|
|
Joely Brammer-Depuy |
joelybd | bu_edu |
Undergraduate researcher |
|
|||
|
|
Alex Seibel |
aseibel | bu_edu |
Postdoctoral fellow Ph.D.
thesis: "Tissue-engineered human
lymphatic models for the study of breast cancer and obesity" |
|
|||
|
|
Madison Tuck |
mtuck | bu_edu |
Undergraduate researcher |
|
|||
|
|
Chitrangada Acharya (2010P) |
|
Research scientist, Allied
Innovative Systems |
|
|||
|
|
Wajd
Al-Holou (2002U) |
|
M.D.,
University of Michigan, with specialization in neurosurgery (2009) |
|
|||
|
|
Nelson Boland (2013-2015U) |
M.D., Baylor College of
Medicine (2019) |
|
||||
|
|
(2011-2012M) |
M.S. thesis: "Genipin Crosslinked Collagen Microfluidic Scaffolds Form Stable
Microvessels In Vitro Using Human Endothelial Cells" M.D., Albert Einstein
College of Medicine (2016) |
|
||||
|
|
Kenneth
Chrobak (2003-2007D) |
Ph.D.
thesis: "Formation of Perfused
Microvessels In Vitro, and Their Use as Models of Barrier Function" Research manager, Baxter
Healthcare |
|
||||
|
|
Cassandra Chua (2017-2020U) |
Medical student, Boston
University |
|||||
|
(2012U) |
Ph.D., biomedical
engineering, Cornell University (2018), with Larry Bonassar Engineer, 3DBio
Therapeutics |
||||||
|
|
Brent Coisman (2013-2014U) |
Research scientist,
Bluebird Bio |
|||||
|
|
Russell
Condie (2006U) |
|
Doctoral
student in biomedical engineering, University of Utah |
||||
|
|
(2014-2015U) |
Ph.D., biomedical
engineering, Case Western Reserve University (2020), with Stathis
Karathanasis Postdoctoral fellow with
Paula Hammond (MIT) |
|||||
|
|
Yoseph
Dance (2018-2022D) |
Ph.D.
thesis: "Engineering 3D Breast
Tumor-on-a-Chip Devices to Investigate the Roles of Adipose Tissue and
Obesity in the Early Stages of Breast Cancer Metastasis" Postdoctoral
fellow with Jude Phillip (Johns Hopkins) |
|||||
|
|
Hillary
Eggert (2004U) |
|
Carthage
College, Biology |
||||
|
|
Evan Feldman (2014-2015U) |
Software engineer,
Viridis3D |
|||||
|
|
Yixin
Gao (2023-2025U) |
|
M.S.,
biomedical engineering, Boston University (2025) |
|
|||
|
|
Andrew
Golden (2002-2008D) |
Ph.D.
thesis: "Microfluidic Hydrogels
for Microvascular Tissue Engineering" Research manager,
AgaMatrix |
|||||
|
|
John
Jiang (2017-2019S) |
Animal
surgeon, Boas Lab, Boston University |
|||||
|
|
Owen
Kelly (2020-2023U) |
Doctoral
student in biomedical engineering, University of Michigan |
|
||||
|
|
(2011-2013U) |
Doctoral student in
biophysics, UC Santa Barbara |
|||||
|
|
Nikhil
Lahiri (2022-2025U) |
|
B.S.,
biomedical engineering, Boston University (2025) |
|
|||
|
Alex Leung (2009-2011U) |
|
M.D., Boston University,
with specialization in vascular surgery (2016) |
|||||
|
|
Henry
Li (2015-2019D) |
Ph.D.
thesis: "Engineering Components of
a Vascularized, Microsurgically Implantable Adipose Tissue" Research
manager, WuXi Biologics |
|||||
|
|
(2014-2016U) |
Ph.D., biomedical
engineering, Johns Hopkins University (2021), with Peter Searson Postdoctoral fellow with
Myriam Heiman (Broad Institute) |
|||||
|
|
Chao Liu (2017-2019U) |
Doctoral
student in biomedical engineering, Case Western Reserve University |
|||||
|
|
(2014-2018U) |
Ph.D., biomedical
engineering, University of Michigan (2023), with Andy Putnam Research scientist, IVIVA
Medical |
|||||
|
|
Jordann Marinelli (2015-2017U) |
Consultant, Accenture |
|||||
|
Brandon Markway (2002U) |
Ph.D.,
biomedical engineering, Oregon Health and Science University (2010), with
Owen McCarty and Monica Hinds Research scientist,
Aronora |
||||||
|
|
Miles Massidda (2017-2018U) |
Doctoral student in
biomedical engineering, UT Austin |
|||||
|
|
Cate
McCullough (2003U) |
|
M.S.,
bioengineering, Stanford University (2007) Manager, Durango Machining
Innovations |
||||
|
|
(2015-2018U) |
Doctoral student in
biomedical engineering, Case Western Reserve University |
|||||
|
|
Mackenzie
Obenreder (2020-2022U) |
Doctoral
student in chemical and biological engineering, CU Boulder |
|||||
|
|
Neil
Parikh (2018-2020U) |
|
Medical
student, Boston University |
||||
|
|
Gavrielle
Price (2004-2009D) |
|
Ph.D.
thesis: "Mechanical and Chemical
Control of Barrier in Engineered Microvessels" Science writer, Ironwood
Pharmaceuticals |
||||
|
|
Jason Pui (2011-2012U) |
|
Engineer, Accellent |
||||
|
|
Rachel Roesch (2011U) |
|
Ph.D.,
chemistry, University of Pennsylvania (2017), with Feng Gai Research
scientist, Illumina |
||||
|
|
(2014-2017U) |
|
Medical
student, Boston University School of Medicine |
||||
|
(2008U) |
Ph.D., biomedical
engineering, UT Austin (2016), with Nicholas Peppas Scientist, Dow Corning |
||||||
|
|
(2002-2005D) |
|
Ph.D.
thesis: "In Vitro Engineering of a
Microvascular Network" Assistant professor of
pediatrics, University of Connecticut Health Center |
||||
|
|
Rebecca Thompson (2012-2015U) |
|
Research scientist, Broad
Institute |
||||
|
|
Abed
Tlemcani (2022-2025U) |
B.S.,
biomedical engineering, Boston University (2025) |
|
||||
|
|
James Truslow (2006-2008M; 2008-2011D;
2011-2013P) |
Ph.D. thesis: "Design and Analysis of Engineered
Microvasculature via Computational Methods" M.S. thesis: "Drainage Systems That Maintain
Transmural Pressure in Engineered Microvascular Tissue" Research scientist,
Brigham & Women's Hospital |
|||||
|
|
Kim
Waller (2007-2008U) |
|
Ph.D.,
biomedical engineering, Brown University (2013), with Gregory Jay Program manager, Ximedica |
||||
|
|
(2007-2012D; 2012P) |
Ph.D. thesis: "Normalization of Microvascular
Physiology in Engineered Microvessels via Cyclic Adenosine Monophosphate
Supplementation and Artificial Lymphatic Drainage" Research scientist,
Catamaran Bio |
|||||
|
|
Jingyi Xia (2017-2019U) |
Doctoral student in
biomedical engineering, University of Michigan |
|||||
|
|
Jing Xu (2014-2016U; 2016-2018S) |
M.D., University of
Massachusetts, with specialization in plastic surgery (2023) |
|||||
(P: postdoctoral fellow; D:
doctoral student; M: Master's student; U: undergraduate
researcher; S: research staff)
PUBLICATIONS
71. Seibel, A.J., Frosti, C., Tlemçani, A.R.,
Lahiri, N., Brammer-DePuy, J.A., Layne, M.D. & Tien, J., Obesity-associated
conditions hinder solute drainage function of engineered human lymphatic
vessels. Cell. Mol. Bioeng. 18, 53-69 (2025). [PDF + Supporting Information]
70. Leggett, S.E., Brennan, M.C., Martinez,
S., Tien, J. & Nelson, C.M., Relatively rare populations of invasive cells
drive progression of heterogeneous tumors. Cell.
Mol. Bioeng. 17, 7-24 (2024). [PDF + Supporting Information]
69. Tien, J. & Dance, Y.W. Protein-based
microfluidic models for biomedical applications. In Handbook of the Extracellular Matrix: Biologically-Derived Materials
(eds. Maia, F.R., Reis, R.L. & Oliveira, J.M.), pp. 329-355 (Springer,
Berlin, 2024). [PDF]
68. Dance, Y.W., Obenreder, M.C., Seibel,
A.J., Meshulam, T., Ogony, J.W., Lahiri, N., Pacheco-Spann, L., Radisky, D.C.,
Layne, M.D., Farmer, S.R., Nelson, C.M. & Tien, J., Adipose cells induce
escape from an engineered human breast microtumor independently of their
obesity status. Cell. Mol. Bioeng. 16, 23-39 (2023). [PDF + Supporting Information]
67. Seibel, A.J., Kelly, O.M., Dance, Y.W.,
Nelson, C.M. & Tien, J., Role of lymphatic endothelium in vascular escape
of engineered human breast microtumors. Cell.
Mol. Bioeng. 15, 553-569 (2022).
[PDF + Supporting Information]
66. Dance, Y.W., Meshulam, T., Seibel, A.J.,
Obenreder, M.C., Layne, M.D., Nelson, C.M. & Tien, J., Adipose stroma
accelerates the invasion and escape of human breast cancer cells from an
engineered microtumor. Cell. Mol. Bioeng.
15, 15-29 (2022). [PDF + Supporting Information]
65. Tien, J. & Ghani, U. Methods for
forming human lymphatic microvessels in vitro and assessing their drainage
function. In Biomedical Engineering
Technologies, Volume 2 (Methods in Molecular Biology, vol. 2394) (eds.
Rasooly, A., Baker, H. & Ossandon, M.R.), pp. 651-668 (Humana Press,
Totowa, NJ, 2022). [PDF]
64. Tien, J., Dance, Y.W., Ghani, U., Seibel,
A.J. & Nelson, C.M., Interstitial hypertension suppresses escape of human
breast tumor cells via convection of interstitial fluid. Cell. Mol. Bioeng. 14,
147-159 (2021). [PDF + Supporting
Information]
63. Tien, J. & Dance, Y.W., Microfluidic
biomaterials. Adv. Healthcare Mater. 10, 2001028 (2021). [PDF]
62. Rabie, E.M., Zhang, S.X., Kourouklis,
A.P., Kilinc, A.N., Simi, A.K., Radisky, D.C., Tien, J. & Nelson, C.M.,
Matrix degradation and cell proliferation are coupled to promote invasion and
escape from an engineered human breast microtumor. Integr. Biol. 13, 17-29
(2021). [PDF + Supporting Information]
61. Tien, J., Li, X., Linville, R.M. &
Feldman, E.J., Comparison of blind deconvolution- and Patlak analysis-based
methods for determining vascular permeability. Microvasc. Res. 133,
104102 (2021). [PDF] [Supporting
Information]
60. Tien, J., Ghani, U., Dance, Y.W., Seibel,
A.J., Karakan, M.C., Ekinci, K.L. & Nelson, C.M., Matrix pore size governs
escape of human breast cancer cells from a microtumor to an empty cavity. iScience 23,
101673 (2020). [PDF + Supporting
Information]
59. Li, X., Xu, J., Bartolák-Suki, E., Jiang, J.
& Tien, J., Evaluation of 1-mm-diameter endothelialized dense collagen
tubes in vascular microsurgery. J. Biomed. Mater. Res. B 108,
2441-2449 (2020). [PDF]
58. Tien, J., Tissue engineering of the
microvasculature. Compr. Physiol. 9, 1155-1212 (2019). [PDF]
57. Li,
X., Xia, J., Nicolescu, C.T., Massidda, M.W., Ryan, T.J. & Tien, J.,
Engineering of microscale vascularized fat that responds to perfusion with
lipoactive hormones. Biofabrication 11, 014101 (2019). [PDF]
56. Thompson, R.L., Margolis, E.A.,
Ryan, T.J., Coisman, B.J., Price, G.M., Wong, K.H.K. & Tien, J., Design
principles for lymphatic drainage of fluid and solutes from collagen scaffolds.
J. Biomed. Mater. Res. A 106,
106-114 (2018). [PDF]
55. Li,
X., Xu, J., Nicolescu, C.T., Marinelli, J.T. & Tien, J., Generation,
endothelialization, and microsurgical suture anastomosis of strong
1-mm-diameter collagen tubes. Tissue Eng. A 23, 335-344 (2017). [PDF]
54. Piotrowski-Daspit,
A.S., Simi, A.K., Pang, M.-F., Tien, J. & Nelson, C.M. A three-dimensional
culture model to study how fluid pressure and flow affect the behavior of
aggregates of epithelial cells. In Mammary Gland Development (Methods in
Molecular Biology, vol. 1501) (eds. Martin, F., Stein, T. & Howlin,
J.), pp. 245-257 (Humana Press, New York, NY, 2017). [PDF]
53. Piotrowski-Daspit, A.S., Tien, J. &
Nelson, C.M., Interstitial fluid pressure regulates collective invasion in
engineered human breast tumors via
Snail, vimentin, and E-cadherin. Integr. Biol. 8, 319-331 (2016). [PDF + Supporting Information]
52. Linville,
R.M., Boland, N.F., Covarrubias, G., Price, G.M. & Tien, J., Physical and
chemical signals that promote vascularization of capillary-scale channels. Cell.
Mol. Bioeng. 9, 73-84 (2016). [PDF]
51. Tien,
J., Li, L., Ozsun, O. & Ekinci, K.L., Dynamics of interstitial fluid
pressure in extracellular matrix hydrogels in microfluidic devices. J.
Biomech. Eng. 137, 091009 (2015). [PDF]
50. Ozsun, O.,
Thompson, R.L., Ekinci, K.L. & Tien, J., Non-invasive mapping of
interstitial fluid pressure in microscale tissues. Integr. Biol. 6,
979-987 (2014). [PDF]
49. Tien, J., Microfluidic approaches for
engineering vasculature. Curr. Opin. Chem. Eng. 3, 36-41 (2014). [PDF]
48. Chan, K.L.S., Khankhel, A.H., Thompson,
R.L., Coisman, B.J., Wong, K.H.K., Truslow, J.G. & Tien, J., Crosslinking
of collagen scaffolds promotes blood and lymphatic vascular stability. J.
Biomed. Mater. Res. A 102,
3186-3195 (2014). [PDF]
47. Tien, J. & Nelson, C.M.,
Microstructured extracellular matrices in tissue engineering and development,
an update. Ann. Biomed. Eng. 42, 1413-1423 (2014). [PDF]
46. Wong, K.H.K., Truslow, J.G., Khankhel,
A.H. & Tien, J. Biophysical mechanisms that govern the vascularization of
microfluidic scaffolds. In Vascularization: Regenerative Medicine and Tissue
Engineering (ed. Brey, E.M.), pp. 109-124 (CRC Press, Boca Raton, FL,
2014). [PDF]
45. Truslow, J.G. & Tien, J.,
Determination of vascular permeability coefficients under slow lumenal filling.
Microvasc. Res. 90, 117-120 (2013). [PDF]
44. Wong, K.H.K., Truslow, J.G., Khankhel,
A.H., Chan, K.L.S. & Tien, J., Artificial lymphatic drainage systems for
vascularized microfluidic scaffolds. J. Biomed. Mater. Res. A 101,
2181-2190 (2013). [PDF]
43.
Tien, J., Wong, K.H.K. & Truslow, J.G. Vascularization of microfluidic
hydrogels. In Microfluidic Cell Culture Systems (eds. Bettinger, C.J.,
Borenstein, J.T. & Tao, S.L.), pp. 205-221 (Elsevier, Oxford, U.K., 2013). [PDF]
42. Tien, J., Truslow, J.G. & Nelson,
C.M., Modulation of invasive phenotype by interstitial pressure-driven
convection in aggregates of human breast cancer cells. PLoS One 7, e45191 (2012). [Corrected PDF + Supporting Information]
41. Leung, A.D., Wong, K.H.K. & Tien, J.,
Plasma expanders stabilize human microvessels in microfluidic scaffolds. J.
Biomed. Mater. Res. A 100, 1815–1822 (2012). [PDF]
40.
Wong, K.H.K., Chan, J.M., Kamm, R.D. & Tien, J., Microfluidic models of
vascular functions. Annu. Rev. Biomed. Eng. 14,
205–230 (2012). [PDF]
39.
Truslow, J.G. & Tien, J., Perfusion systems that minimize vascular volume
fraction in engineered tissues. Biomicrofluidics 5, 022201 (2011). [PDF]
38. Price, G.M. & Tien, J. Methods
for forming human microvascular tubes in vitro and measuring their
macromolecular permeability. In Biological Microarrays (Methods in Molecular
Biology, vol. 671) (eds. Khademhosseini, A., Suh, K.-Y. & Zourob, M.),
pp. 281-293 (Humana Press, Totowa, NJ, 2011). [PDF]
37. Price, G.M., Wong, K.H.K., Truslow,
J.G., Leung, A.D., Acharya, C. & Tien, J., Effect of mechanical factors on
the function of engineered human blood microvessels in microfluidic collagen
gels. Biomaterials 31,
6182-6189 (2010). [PDF]
36. Wong, K.H.K., Truslow, J.G. &
Tien, J., The role of cyclic AMP in normalizing the function of
engineered human blood microvessels in microfluidic collagen gels. Biomaterials 31, 4706-4714 (2010). [PDF] [Movie]
35. Truslow, J.G., Price, G.M. &
Tien, J., Computational design of drainage systems for vascularized scaffolds. Biomaterials
30, 4435-4443 (2009).
[PDF]
34. Price,
G.M. & Tien, J. Subtractive methods for forming microfluidic gels of
extracellular matrix proteins. In Microdevices in Biology and Engineering
(eds. Bhatia, S.N. & Nahmias, Y.), pp. 235-248 (Artech House, Boston, MA, 2009). [PDF]
33. Price, G.M., Chu, K.K., Truslow, J.G.,
Tang-Schomer, M.D., Golden, A.P., Mertz, J. & Tien, J., Bonding of
macromolecular hydrogels using perturbants. J.
Am. Chem. Soc. 130,
6664-6665 (2008). [PDF + Supporting Information]
[Movies]
32. Price, G.M., Chrobak, K.M. & Tien,
J., Effect of cyclic AMP on barrier function of human lymphatic microvascular
tubes. Microvasc. Res. 76, 46-51 (2008). [PDF]
31. Golden, A.P. & Tien, J., Fabrication
of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab
Chip 17, 720-725 (2007). [PDF]
30. Nelson, C.M. & Tien, J., Microstructured
extracellular matrices in tissue engineering and development. Curr. Opin. Biotechnol. 17, 518-523 (2006). [PDF]
29. Chrobak, K.M., Potter, D.R. & Tien,
J., Formation of perfused, functional microvascular tubes in vitro. Microvasc. Res. 71, 185-196 (2006). [PDF] [Movies]
28. Tien, J., Golden, A.P. & Tang, M.D.
Engineering of blood vessels. In Microvascular
Research: Biology and Pathology, Vol. 2 (eds. Shepro, D. & D'Amore,
P.A.), pp. 1087-1093 (Elsevier Academic Press, San Diego, CA, 2006). [PDF]
27. Tang, M.D., Golden, A.P. & Tien, J.,
Fabrication of collagen gels that contain patterned, micrometer-scale cavities.
Adv. Mater. 16, 1345-1348 (2004). [PDF]
26. Gray, D.S., Tien, J. & Chen, C.S.,
High conductivity elastomeric electronics. Adv.
Mater. 16, 393-397 (2004). [PDF]
25. Chen, C.S., Tan, J.L. & Tien, J.,
Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 6,
275-302 (2004). [PDF]
24. Tang, M.D., Golden, A.P. & Tien, J.,
Molding of three-dimensional microstructures of gels. J. Am. Chem. Soc. 125,
12988-12989 (2003). [PDF]
23. Gray, D.S., Tien, J. & Chen, C.S.,
Repositioning of cells by mechanotaxis on surfaces with micropatterned Young's
modulus. J. Biomed. Mater. Res. A 66, 605-614 (2003). [PDF]
22. Tan, J.L., Tien, J., Pirone, D.M., Gray,
D.S., Bhadriraju, K. & Chen, C.S., Cells lying on a bed of microneedles: an
approach to isolate mechanical force. Proc.
Natl. Acad. Sci. USA 100,
1484-1489 (2003). [PDF]
21. Clark, T.D., Ferigno, R., Tien, J., Paul,
K.E. & Whitesides, G.M., Template-directed self-assembly of
10-μm-sized hexagonal plates. J. Am.
Chem. Soc. 124, 5419-5426
(2002). [PDF]
20. Tien, J., Nelson, C.M. & Chen, C.S.,
Fabrication of aligned microstructures with a single elastomeric stamp. Proc. Natl. Acad. Sci. USA 99, 1758-1762 (2002). [PDF]
19. Tien, J. & Chen, C.S., Patterning the
cellular microenvironment. IEEE Eng. Med.
Biol. 21, 95-98 (2002). [PDF]
18. Tan, J.L., Tien, J. & Chen, C.S.,
Microcontact printing of proteins on mixed self-assembled monolayers. Langmuir 18, 519-523 (2002). [PDF]
17. Tien, J. & Chen, C.S. Microarrays of
cells. In Methods of Tissue Engineering
(eds. Atala, A. & Lanza, R.), pp. 113-120 (Academic Press, San Diego, CA,
2001).
16. Bowden, N., Tien, J., Huck, W.T.S. &
Whitesides, G.M. Mesoscale self-assembly: the assembly of micron- and millimeter-sized
objects using capillary forces. In Supramolecular
Organization and Materials Design (eds. Jones, W. & Rao, C.N.R.), pp.
103-145 (Cambridge University Press, New York, NY, 2001).
15. Clark, T.D., Tien, J., Duffy, D.C., Paul,
K.E. & Whitesides, G.M., Self-assembly of 10-μm-sized objects into
ordered three-dimensional arrays. J. Am.
Chem. Soc. 123, 7677-7682
(2001). [PDF]
14. Gracias, D.H., Tien, J., Breen, T.L.,
Hsu, C. & Whitesides, G.M., Forming electrical networks in three dimensions
by self-assembly. Science 289, 1170-1172 (2000). [PDF]
13. Dike, L.E., Chen, C.S., Mrksich, M.,
Tien, J., Whitesides, G.M. & Ingber, D.E., Geometric control of switching
between growth, apoptosis, and differentiation during angiogenesis using
micropatterned substrates. In Vitro Cell.
Dev. Biol. Anim. 35, 441-448
(1999). [PDF]
12. Deng, T., Tien, J., Xu, B. &
Whitesides, G.M., Using patterns in microfiche as photomasks in
10-μm-scale microfabrication. Langmuir
15, 6575-6581 (1999). [PDF]
11. Breen, T.L., Tien, J., Oliver, S.R.J.,
Hadzic, T. & Whitesides, G.M., Design and self-assembly of open, regular,
3D mesostructures. Science 284, 948-951 (1999). [PDF]
10. Lahiri, J., Isaacs, L., Tien, J. &
Whitesides, G.M., A strategy for the generation of surfaces presenting ligands for
studies of binding based on an active ester as a common reactive intermediate. Anal. Chem. 71, 777-790 (1999). [PDF]
9. Tien, J., Breen, T.L. & Whitesides,
G.M., Crystallization of millimeter-scale objects with use of capillary forces.
J. Am. Chem. Soc. 120, 12670-12671 (1998). [PDF]
8. Huck, W.T.S., Tien, J. & Whitesides,
G.M., Three-dimensional mesoscale self-assembly. J. Am. Chem. Soc. 120,
8267-8268 (1998). [PDF]
7. Marzolin, C., Terfort, A., Tien, J. &
Whitesides, G.M., Patterning of a polysiloxane precursor to silicate glasses by
microcontact printing. Thin Solid Films 315, 9-12 (1998). [PDF]
6. Tien, J., Xia, Y. & Whitesides, G.M. Microcontact
printing of SAMs. In Self-Assembled Monolayers of Thiols (Thin Films, vol.
24) (ed. Ulman, A.), pp. 227-254 (Academic Press, San Diego, CA, 1998).
5. Xia, Y., Venkateswaran, N., Qin, D., Tien,
J. & Whitesides, G.M., Use of electroless silver as the substrate in
microcontact printing of alkanethiols and its application in microfabrication. Langmuir 14, 363-371 (1998). [PDF]
4. Mrksich, M., Dike, L.E., Tien, J., Ingber,
D.E. & Whitesides, G.M., Using microcontact printing to pattern the
attachment of mammalian cells to self-assembled monolayers of alkanethiolates
on transparent films of gold and silver. Exp.
Cell Res. 235, 305-313 (1997). [PDF]
3. Tien, J., Terfort, A. & Whitesides,
G.M., Microfabrication through electrostatic self-assembly. Langmuir 13, 5349-5355 (1997). [PDF]
2. Xia, Y., Tien, J., Qin, D. &
Whitesides, G.M., Non-photolithographic methods for fabrication of elastomeric
stamps for use in microcontact printing. Langmuir
12, 4033-4038 (1996). [PDF]
1. Shaw, G.L. & Tien, J., Energy levels
of quark atoms. Phys. Rev. D 47, 5075-5078 (1993). [PDF]
FUNDING
Engineering Vascularized
Models of Obesity Progression (BU Dean’s Catalyst Award)
Persufflation of Composite Tissue Transplants (DoD/Army W81XWH-17-1-0571)
(Re)vascularization of
Decellularized Scaffolds (NIH/NIBIB R03 EB024660)
Engineered
Invasive Human Breast Tumors with Integrated Capillaries and Lymphatics
(NIH/NCI U01 CA214292)
In Vivo Microsurgical
Anastomosis of Prevascularized Tissues (NIH/NIBIB R03 EB018851)
Development and Clinical
Validation of Algorithms for Non-Invasive Mapping of Vascular Permeability
(BU/BWH Partnership Program)
Non-Invasive Measurement of
Vascular Cell Adhesion to Biomaterials (BU Dean’s Catalyst Award)
Active Biomaterials (BU
Materials Science and Engineering Innovation Grant)
Effect
of Interstitial Pressure on Epithelial Invasion from Human Mammary Ducts
(DoD/Army W81XWH-09-1-0565)
Engineering
Functional Lymphatic Networks In Vitro (NIH/NHLBI R21 HL092335)
Synthesis and Characterization of Patterned
Microvascular Networks (NIH/NIBIB R01 EB005792)
Self-Assembly of Mesostructured Biomaterials
(NIH/NIBIB R21 EB003157)
In Vitro Synthesis of a Microvascular Network
(NIH/NIBIB R21 EB002228)
Use of Microfabrication and Self-Assembly in Tissue
Engineering (Whitaker Foundation RG-02-0344)
Dynamic Substrates for Cell Culture (BU Special
Program for Research Initiation Grants)
Self-Assembly of Gels (BU Provost’s Innovation Fund)
Response of Endothelial Cells to Cell-Cell Contact
(NIH/NHLBI F32 HL010486)
LINKS
How to join our research program:
·
Postdoctoral
fellows:
Interested postdoctoral
candidates should send us a detailed cover letter, CV, and a list of three
professional references. We look for
candidates with a robust track record of publication and innovation.
·
Graduate
students:
Graduate students must apply
through one of the doctoral programs listed below—we are especially interested
in applicants with a strong quantitative background and excellent technical
skills:
Department
of Biomedical Engineering
Program in Molecular Biology, Cell Biology, and Biochemistry
Division
of Materials Science and Engineering
Late Entry Accelerated
Program (LEAP) in Biomedical Engineering
MD/PhD program at Boston University
School of Medicine
·
Undergraduate
students:
Undergraduate students
should send us a brief explanatory letter, transcript, and description of any
prior research experience. We seek
students who learn quickly, work hard, and have impeccable ethics.
Resources at BU:
Core facilities (lithography, imaging, and
materials characterization) in the Department of Biomedical Engineering
Core facilities (flow
cytometry, microarrays, transgenics, etc.) at the Medical Center
Core facilities
(lithography, SEM) in the Photonics Center
Collaborators:
Celeste Nelson, Department of
Chemical Engineering, Princeton University
Kamil Ekinci, Department of Mechanical
Engineering, Boston University
Matt Layne,
Department of Biochemistry, Boston University School of Medicine
Databases and analytical software:
ISI Web of Knowledge (here,
for BU users)
The Lipid Library (with focus on
bioactive lipids)
Atlas of microsurgery
Statistical tests and when to use them
Don't know where to
publish? Ask JANE!
Journals of particular relevance to
microcirculation:
American Journal of
Physiology – Heart and Circulatory Physiology
Journal of Experimental
Medicine
Lymphatic Research and Biology
Organizations:
National Institutes of Health (NIH)
National Institute of Biomedical Imaging and
Bioengineering (NIBIB)
National Heart, Lung, and Blood Institute
(NHLBI)
National Cancer Institute (NCI)
Information
on funded NIH grants: RePORTER database
Information
on deadlines,
study
sections, special
emphasis panels, funding strategies,
and opportunities
National Science Foundation (NSF)
Biomedical Engineering Society (BMES)
American Heart Association (AHA)
Lymphatic Education and Research Network
Organ Procurement and Transplantation
Network (OPTN)
NHS Blood and Transplant (NHSBT)
World
Marrow Donor Association
Upcoming events:
Seminars at BU in biomedical
engineering and systems
biology
Courses
at the Marine Biology Laboratory/Woods Hole
Courses at Cold Spring Harbor
Laboratory
BMES 2025 Annual Meeting
(Oct 8-11, 2025; San Diego, CA)
[Copyright © 2025 by the
Tien Group.]




































